|
|
| Line 1: |
Line 1: |
| | + | This page is under construction, Will be completed by 6/30 |
| | + | Following this procedure a user will be able to submit jobs to lochness or stheno from Matlab running locally on the user's computer. The version of Matlab on the user's computer must be the same as on the cluster, currently 2021a. |
| | + | ==Installing the Add-On== |
| | | | |
| − | ==<strong><font color="#9966cc">Accessing AFS Space</font></strong>==
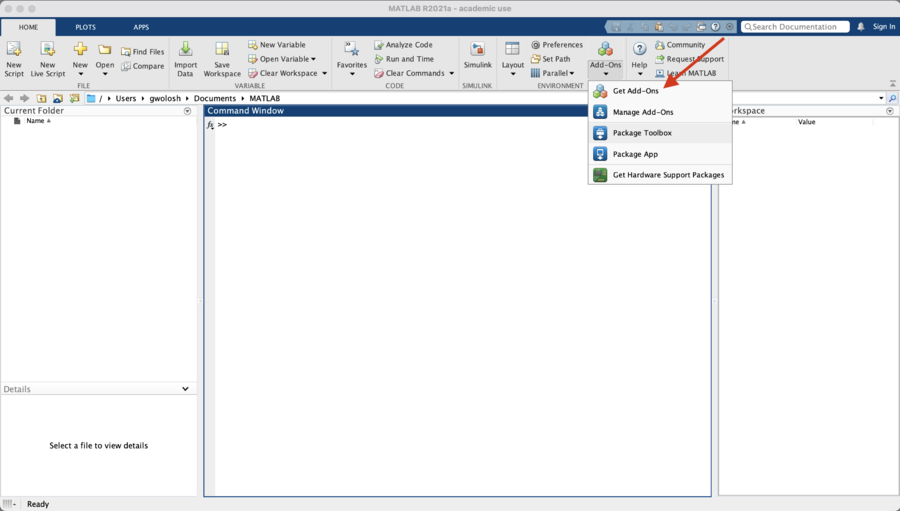
| + | From the Matlab window, click on "Add-ons" and select "Get Add-Ons." |
| − | ===How can I access my AFS space from HPC cluster compute nodes ?===
| + | |
| − | Jobs submitted by the SGE scheduler to compute nodes have to get the user's
| + | |
| − | AFS token in order to access space in AFS that that user has access to.
| + | |
| − | The current method for doing this is via "ksub". See [[UsingKsub]].
| + | |
| | | | |
| − | Just before running <em>ksub</em>, be <strong>sure</strong> you have your Kerberos
| + | [[File:ClickOnAddons.png|900px]] |
| − | ticket and AFS token by :
| + | |
| − | <pre code>kinit && aklog</pre>
| + | |
| | | | |
| − | An improved method for doing what ksub does, "auks", is in the process of being implemented.
| + | In the search box enter "slurm" and click on the magnifying glass icon. |
| | | | |
| − | ==<strong><font color="#9966cc">Compilers</font></strong>==
| + | Select "Parallel Computing Toolbox plugin for MATLAB Parallel Server with Slurm" |
| − | ===What compilers are available ?===
| + | |
| − | In addition to the compilers listed by "module av", the GNU compilers that are
| + | |
| − | part of the standard operating system installation are available.
| + | |
| | | | |
| − | See [[SoftwareModulesAvailable]]
| + | Alternatively, this Add-On can be downloaded directly from the [https://www.mathworks.com/matlabcentral/fileexchange/52807-parallel-computing-toolbox-plugin-for-matlab-parallel-server-with-slurm Mathworks] site. |
| | | | |
| − | ==<strong><font color="#9966cc">Compiling CUDA code</font></strong>==
| + | [[File:SlurmAddOn.png|1000px]] |
| − | ===How do I compile CUDA programs ?===
| + | |
| − | <ol>
| + | |
| − | <li>
| + | |
| − | Get an interactive login a GPU node, e,g., node151 or node152
| + | |
| − | <pre code>qlogin node151</pre>
| + | |
| − | </li>
| + | |
| | | | |
| − | <li>
| + | Click on "Install." |
| − | Load gcc and CUDA modules, e.g.,
| + | |
| − | <pre code>module load gcc/5.4.0 cuda</pre>
| + | |
| − | </li>
| + | |
| | | | |
| − | <li>
| + | [[File:ClickOnInstall.png|1000px]] |
| − | Compile your code
| + | |
| − | </li>
| + | |
| | | | |
| − | <li>
| + | The installation of the Add-On is complete. Click on "OK" the start the "Generic Profile Wizard for Slurm." |
| − | Log out of the GPU node
| + | <br> |
| − | </li>
| + | |
| | | | |
| − | <li>
| + | [[File:InstallationComplete.png|900px]] |
| − | Submit your job to the <strong>gpu</strong> queue using a submit script
| + | ==Creating a Profile for Lochness or Stheno== |
| − | </li>
| + | |
| − | </ol>
| + | |
| | | | |
| − | See [[RunningCUDASamplesOnKong]] and [[KongQueuesTable]]
| + | The following steps will create a profile for lochness (or stheno). |
| | + | Click "Next" to begin. |
| | | | |
| − | ==<strong><font color="#9966cc">Conserving disk space</font></strong>==
| + | [[File:GenericProfile1.png|800px]] |
| − | ===What are the best ways of conserving local space on the HPC clusters ?===
| + | |
| − | <ol>
| + | |
| − | <li>Remove unnecessary files</li>
| + | |
| − | <li>If you have a research directory located at /afs/cad/research/..,
| + | |
| − | move (and better yet compress) files into a sub-directory of that
| + | |
| − | directory. This not only frees up space on on the cluster, but keeps the
| + | |
| − | user's results available even after their cluster account is removed.
| + | |
| − | </li>
| + | |
| − | <li>Compress files with <em>bzip2</em> or <em>gzip</em>. bzip2 is usually more effective</li>
| + | |
| − | </ol>
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">"Error message: vsu_ClientInit: Could not get afs tokens, running unauthenticated" from qsub</font></strong>==
| + | In the "Operating System" screen "Unix" is already selected. Click "Next" to continue. |
| − | ===I get this message when running <em>qsub</em>. Is it something I should be concerned about ?===
| + | |
| − | This message indicates that you do not have your AFS token. The message can be safely
| + | |
| − | ignored when running qsub.
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Error message from a node</font></strong>==
| + | [[File:GenericProfile2.png|800px]] |
| − | ===I get this message repeatedly in a terminal window when logged into Kong. Is it something I should be concerned about ?===
| + | |
| − | This message indicates that there is a problem with a certain node. The message can be safely ignored, unless your job was
| + | |
| − | using that node. The ARCS staff will take the offending node out of service as soon as possible.
| + | |
| − | <pre code>
| + | |
| − | Message from syslogd@nodeXXX at timestamp
| + | |
| − | kernel: Code: 48 8d 45 d0 4c 89 4d f8 c7 45 b0 10 00 00 00 48 89 45 c0 e8 38 ff ff ff c9 c3 90 90 90 90 90 90 44 8d 46 3f 85 f6 55 44
| + | |
| − | 0f 49 c6 <31> d2 48 89 e5 41 c1 f8 06 45 85 c0 7e 24 48 83 3f 00 48 89 f8
| + | |
| − | </pre>
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Getting access to HPC clusters</font></strong>==
| + | This "Submission Mode" screen determines whether or not to use a "shared" or "nonshared" submission mode. Since Matlab installed on your personal computer or laptop does not use a shared job location storage, select "No" where indicated and click "Next" to continue. |
| − | ===Who is eligible ?===
| + | |
| − | All NJIT researchers; courses using HPC. See [[UserAccess]].
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Getting AFS token using qlogin</font></strong>==
| |
| − | ===How do I get my AFS token when using <em>qlogin</em> ?===
| |
| − | After qlogin :
| |
| − | <pre code>
| |
| − | kinit && aklog && tokens
| |
| − | </pre>
| |
| | | | |
| − | <em>kinit</em> gets your Kerberos ticket when you supply the correct password
| + | [[File:GenericProfile3.png|800px]] |
| | | | |
| − | <em>aklog</em> gets your AFS token from your Kerberos ticket
| + | Click "Next" to continue. |
| | | | |
| − | <em>tokens</em> displays your AFS token status
| + | [[File:GenericProfile4.png|800px]] |
| | | | |
| − | ==<strong><font color="#9966cc"> Kerberos ticket and AFS token status message</font></strong>==
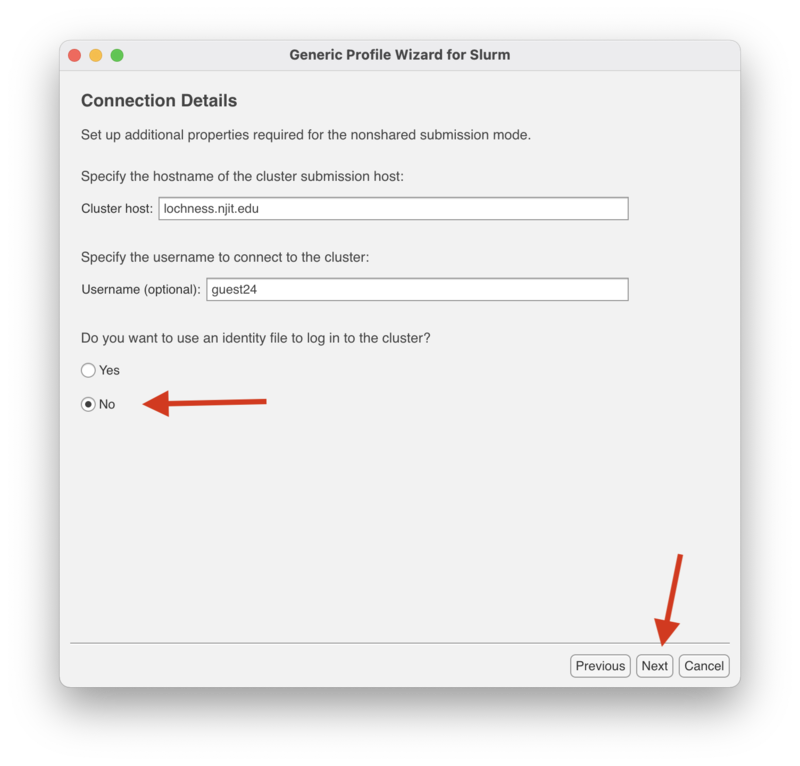
| + | In the "Connection Details" screen, enter the cluster host, either "lochness.njit.edu" or "stheno.njit.edu." |
| − | ===<strong><font color="#9966cc">What does the "Your Kerberos ticket and AFS token status" message mean</font></strong> ?===
| + | Enter your UCID for the username. |
| | | | |
| − | A message such as the following is informational, telling the user :
| + | Select "No" for the "Do you want to use an identity file to log in to the cluster" option and click next to continue. |
| − | <ul>
| + | |
| − | <li>Until what date thier Kerberos tickect ca be renewed</li>
| + | |
| − | <li>When their AFS token will expire, unless refreshed</li>
| + | |
| − | </ul>
| + | |
| | | | |
| − | <pre code>
| + | [[File:GenericProfile5.png|800px]] |
| − | === === === Your Kerberos ticket and AFS token status === === ===
| + | |
| − | Kerberos : Renew until 02/25/17 07:36:21, Flags: FRI
| + | |
| − | AFS : User's (AFS ID 22964) tokens for afs@cad.njit.edu [Expires Jan 31 17:36]
| + | |
| − | </pre>
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Getting cluster resource usage history</font></strong>==
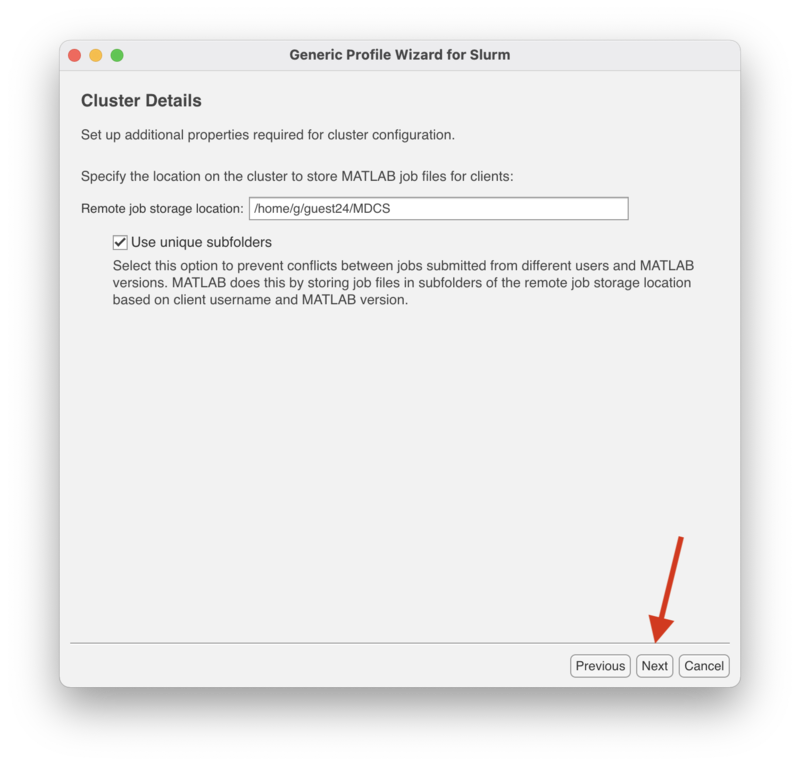
| + | In the "Cluster Details" screen enter the full path to the directory on lochness to store the Matlab job files. In the case the directory is $HOME/MDCS. MDCS stands for Matlab Distributed Computing Server. It is not necessary to name this directory MDCS. This directory can be named anything you wish. To determine the value of $HOME, log onto lochness and run the following: |
| − | ===How can I view the historical usage of resources on the HPC clusters ?===
| + | |
| − | Use [[Ganglia]]
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Getting files into your local directory on kong or stheno</font></strong>==
| + | <pre> |
| − | ===How do I get files from outside kong or stheno into my local directory on those machines ?===
| + | login-1-45 ~ >: echo $HOME |
| − | The easiest way to to this is to copy the files from AFS.
| + | /home/g/guest24</pre> |
| − | <ul>
| + | |
| − | <li>Since kong and stheno are AFS clients, a user logged into kong or stheno can copy files from any location in
| + | |
| − | AFS that the user has read access into their local kong or stheno directory.</li>
| + | |
| | | | |
| − | <li>Conversely, a user can copy files from their local kong or stheno directory to any
| + | Make sure to check the box "Use unique subfolders." |
| − | location in AFS to which the user has write access - e.g., a research directory or AFS home directory.</li>
| + | |
| − | </ul>
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Archiving data</font></strong>==
| + | Click "Next" to continue. |
| − | ===How do I archive data from local storage on a cluster or AFS ?===
| + | |
| − | <ol>
| + | |
| − | <li>Use rclone : [[https://rclone.org/ rclone]]</li>
| + | |
| − | <li>rsync to a local disk - contact <em>arcs@njit.edu</em> for assistance
| + | |
| − | </ol>
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Getting Job Status</font></strong>==
| + | [[File:GenericProfile6.png|800px]] |
| − | ===How can I get the status of jobs I have submitted to SGE ?===
| + | |
| − | Use "qtsat" on a head node in various formats. See [[SonOfGridEngine]].
| + | |
| | | | |
| − | ===How can I see an overall summary of queue activity ?===
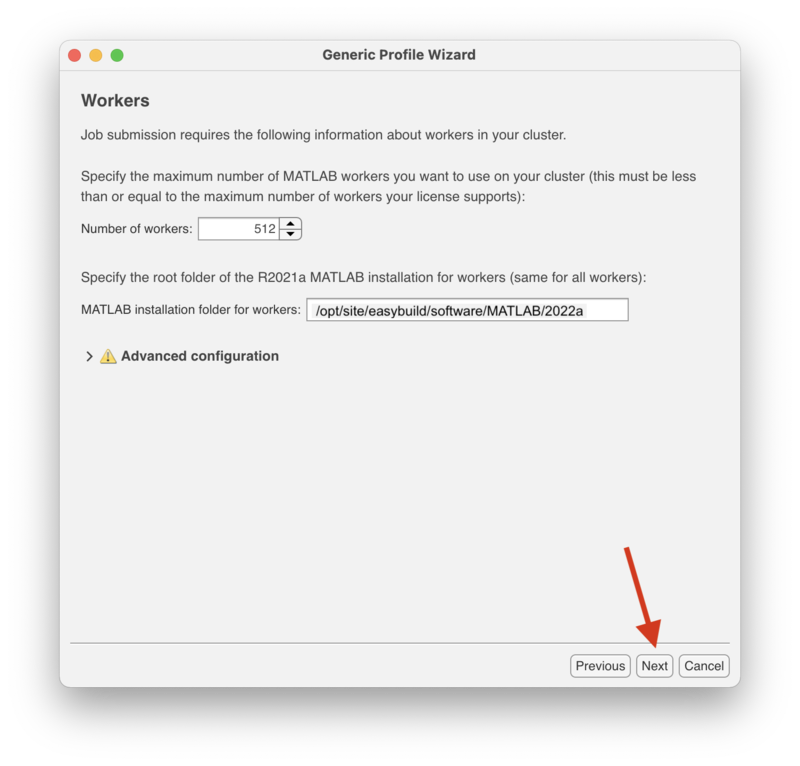
| + | In the "Workers" screen enter "512" for the number of workers and "/opt/site/apps/matlab/R2021a" for "MATLAB installation folders for workers." |
| − | Use "qsummary" on a head node. For usage : "qsummary -h". See [[SonOfGridEngine]].
| + | Click "Next" to continue. |
| | | | |
| − | ==<strong><font color="#9966cc">Getting local files onto or asccessible from an HPC cluster</font></strong>==
| + | [[File:GenericProfile7_1.png|800px]] |
| − | ===How do I get files that are local to my computer onto or accessible from the kong or stheno clusters ?===
| + | |
| − | Programs running on compute nodes can access files that are :
| + | |
| − | <ol>
| + | |
| − | <li>in the user's local cluster home directory</li>
| + | |
| − | <li>in an AFS directory the user has access to</li>
| + | |
| − | </ol>
| + | |
| − | 1. Getting files from your local computer to your local cluster home directory
| + | |
| − | <ol type="A">
| + | |
| − | <li>make your local computer an AFS client; contact <em>arcs@njit.edu</em> for help with this</li>
| + | |
| − | <li>copy the relevant files from your local computer into AFS space that you have write access to,
| + | |
| − | usually a research directory</li>
| + | |
| − | <li>log in to the cluster headnode; make sure you have your AFS token, via <em>tokens</em>; use "kinit
| + | |
| − | && aklog" if you don't have a token</li>
| + | |
| − | <li>use <em>tar</em> or <em>cp</em> to get the files in AFS into your local cluster home
| + | |
| − | directory. Contact <em>arcs@njit.edu</em> if you need help</li>
| + | |
| − | </ol>
| + | |
| − | 2. Accessing your files in AFS directly from compute nodes
| + | |
| − | <ul>
| + | |
| − | <li>Use ksub [[UsingKsub]]</li>
| + | |
| − | </ul>
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Languages</font></strong>==
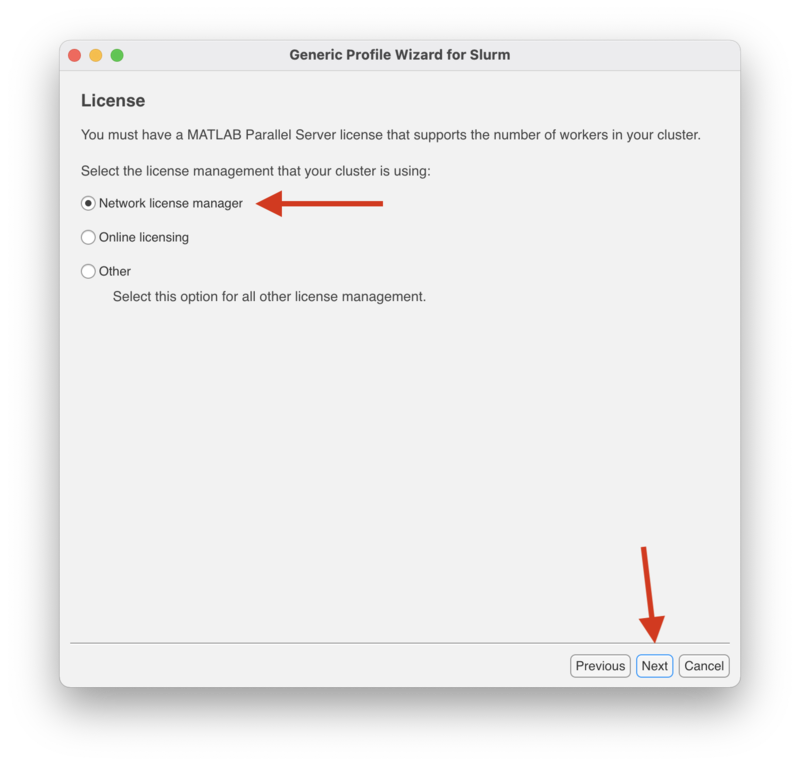
| + | In the "License" screen make sure to select "Network license manager" and click "Next" to continue. |
| − | ===What languages are available ?===
| + | |
| − | In addition to the languages listed by "module av", the languages that are
| + | |
| − | part of the standard operating system installation are available - e.g.,
| + | |
| − | /usr/bin/perl, /usr/bin/python. See [[SoftwareModulesAvailable]].
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Performance, local machine vs. cluster</font></strong>==
| + | [[File:GenericProfile8.png|800px]] |
| − | ===An application runs faster on my own computer than it does on the HPC cluster ...?===
| + | |
| − | This can happen for a variety of reasons, including :
| + | |
| − | <ul>
| + | |
| − | <li>your local computer has faster CPU, more RAM than cluster nodes</li>
| + | |
| − | <li>your job on the cluster node shares the nodes's reosurces with other jobs</li>
| + | |
| − | <li>the disk I/O on your local computer is faster than on the cluster</li>
| + | |
| − | </ul>
| + | |
| − | It should be noted that even in such cases users can get significantly better theoughput using
| + | |
| − | a cluster by :
| + | |
| − | <ul>
| + | |
| − | <li>running many serial jobs simultaneously</li>
| + | |
| − | <li>running jobs in parallel</li>
| + | |
| − | </ul>
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Program runs on head node but not on compute nodes</font></strong>==
| + | In the "Profile Details" screen enter either "Lochness" or "Stheno" depending on which cluster you are making a profile for. The "Cluster description" is optional and may be left blank. |
| − | ===My program runs OK when I run it on the head node, but produces different results, or doesn't run at all, on compute nodes. Why ?===
| + | Click "Next" to continue. |
| − | There remain differences - scheduled to be remedied - between the head node and compute nodes. Code
| + | |
| − | compiled on the head node may not run the same way on compute nodes. Fix : use <em>qlogin</em> to log in
| + | |
| − | to an arbitrary compute node, and compile your code on that node. | + | |
| | | | |
| − | ==<strong><font color="#9966cc">Resources for big data analysis</font></strong>==
| + | [[File:GenericProfile9.png|800px]] |
| − | ===What resources exist for big data analysis ?===
| + | |
| − | An Hadoop cluster for research and teaching came on-line 09 Dec 2015. Documentation on
| + | |
| − | its use is being developed.
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Resources writeups</font></strong>==
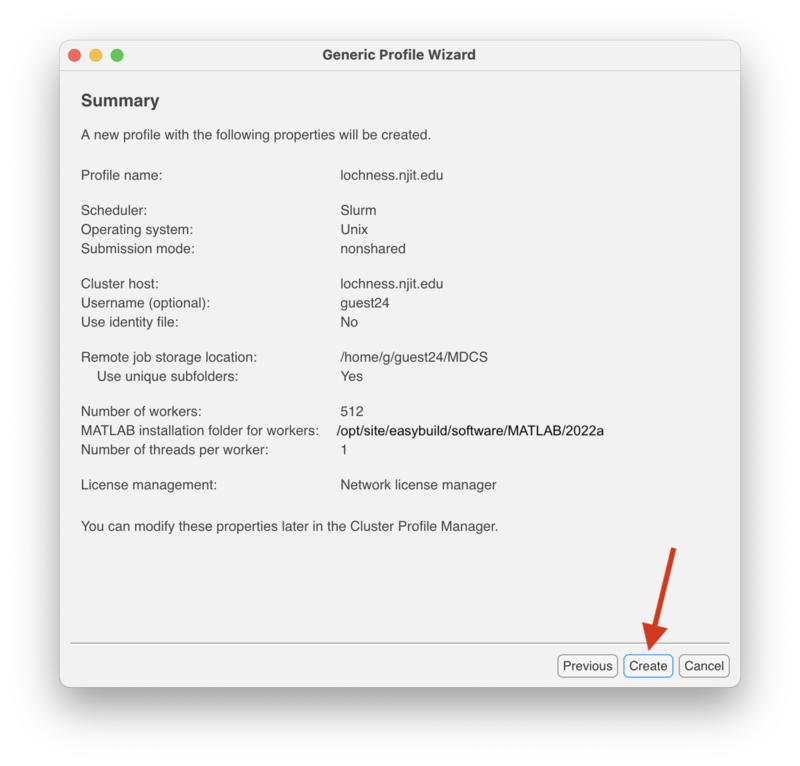
| + | In the "Summary" screen make sure everything is correct and click "Create." |
| − | ===What writeups are available that describe NJIT's high performance computing (HPC) and big data (BD) resources ?===
| + | |
| − | IST support for researchers [[ISTResearcherSupport]]
| + | |
| | | | |
| − | Short Overview [[ShortOverview]]
| + | [[File:GenericProfile10_1.png|800px]] |
| | | | |
| − | Very Short Overview [[VeryShortOverview]]
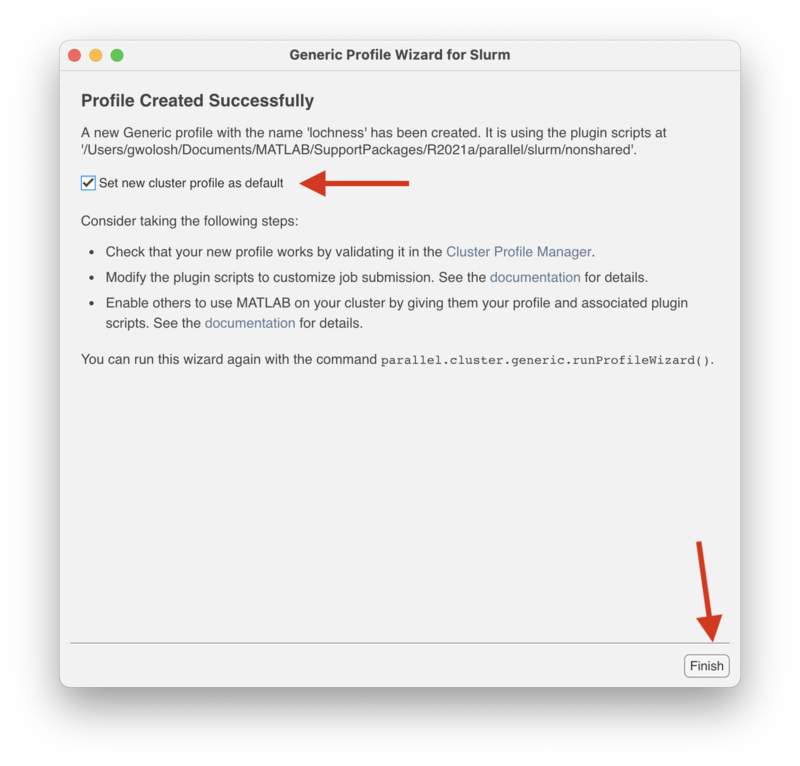
| + | In the "Profile Created Successfully" screen, check the "Set the new profile as default" box and click on "Finish." |
| | | | |
| − | Detailed hardware specs [https://web.njit.edu/topics/hpc/specs Cluster specs]
| + | [[File:GenericProfile11.png|800px]] |
| | | | |
| − | ==<strong><font color="#9966cc">Running Matlab</font></strong>== | + | ==Submitting a Serial Job== |
| − | ===How do I specify matlab jobs to use a specific queue ?===
| + | |
| − | A queue can be specified within a matlab input file, e.g. :
| + | |
| − | <pre code>
| + | |
| − | ClusterInfo.setQueue ('medium')
| + | |
| − | </pre>
| + | |
| | | | |
| − | ===Is there support for Matlab Distributed Computing Srever (MDSC) on the HPC clusters ?===
| + | This section will demonstrate how to create a cluster object and submit a simple job to the cluster. The job will run the 'hostname' command on the node assigned to the job. The output will indicate clearly that the job ran on the cluster and not on the local computer. |
| − | Yes. See [[GettingStartedWithSerialAndParallelMATLABOnKongAndStheno]].
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Software</font></strong>==
| + | The hostname.m file used in this demonstration can be downloaded [https://www.mathworks.com/matlabcentral/fileexchange/24096-hostname-m here.] |
| − | ===What software is available on the HPC clusters ?===
| + | |
| − | The simplest way to see what software is available is to enter : | + | |
| − | <pre code>
| + | |
| − | module available
| + | |
| − | </pre>
| + | |
| − | This will produce a list of most - but not all - available software.
| + | |
| | | | |
| − | You can refine your search to list specific software. E.g., to list
| + | In the Matlab window enter: |
| − | all of the GCC compilers avaialable, :
| + | |
| − | <pre code>
| + | |
| − | module available gcc
| + | |
| − | </pre>
| + | |
| | | | |
| − | Recent list of modules [[SoftwareModulesAvailable]]
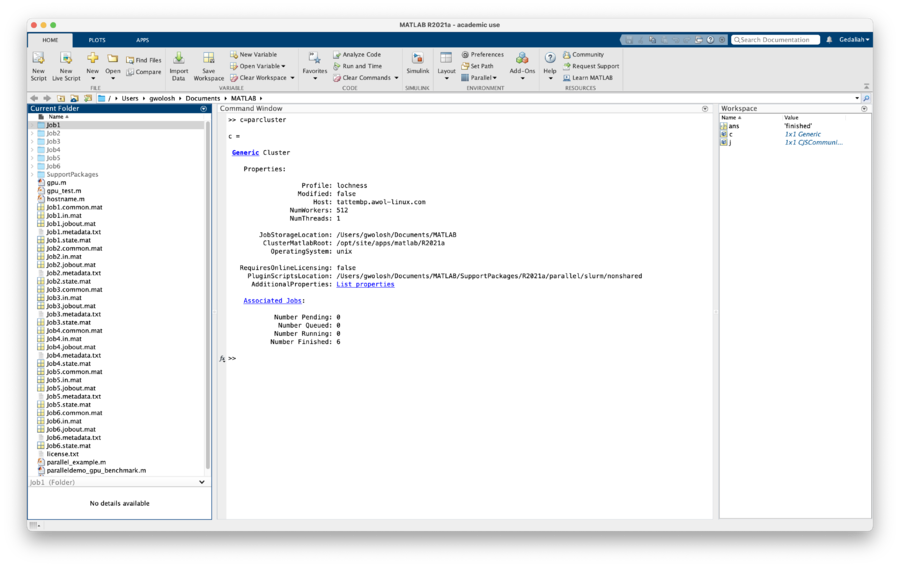
| + | <pre> >> c=parcluster </pre> |
| | | | |
| − | The "module available" command can be used on the kong and stheno
| + | [[File:c=parcluster_1.png|900px]] |
| − | head nodes, and on all public-access AFS Linux clients. The shortest
| + | |
| − | form of this command is "module av".
| + | |
| | | | |
| − | ===What is the "module" Command ?===
| + | Certain arguments need to be passe dto SLURM in order for the job to run properly. Her we will set values for partion, mem-per-cpu and time. In the Matlab window enter: |
| − | This command is used to set the environment variables for the software you
| + | |
| − | want to use. see [https://en.wikipedia.org/wiki/Environment_Modules_(software) Environt modules],
| + | |
| − | and "man module". | + | |
| | | | |
| − | ===How do I use the "module load <software>" command ?===
| + | <pre> >> c.AdditionalProperties.AdditionalSubmitArgs=['--partition=public --mem-per-cpu=10G --time=2-00:00:00'] </pre> |
| − | Generally, this command is placed in your submit script, as in :
| + | |
| − | <pre code>
| + | |
| − | module load matlab
| + | |
| − | </pre> | + | |
| | | | |
| − | ===How do I request software that is not currently available ?===
| + | To make this persistent between Matlab sessions these arguments need to be saved to the profile. In the Matlab window enter: |
| − | This is done by sending mail to <em>arcs@njit.edu</em>. The software must meet these
| + | |
| − | criteria :
| + | |
| − | <ul>
| + | |
| − | <li>requested by faculty or staff</li>
| + | |
| − | <li>free</li>
| + | |
| − | <li>compatible with the current Linux version on the HPC clusters</li>
| + | |
| − | <li>used for research and/or courses</li>
| + | |
| − | </ul>
| + | |
| − | In general, such software will be installed in AFS, and will be accessible to
| + | |
| − | all AFS Linux clients.
| + | |
| | | | |
| − | If the software is not free, a funding source will need to be identified.
| + | <pre> >> c.saveProfile </pre> |
| | | | |
| − | ===Running Jupyter Notebook===
| + | [[File:AdditionalArguments.png|900px]] |
| − | <ul>
| + | |
| − | <li>copy /opt/site/examples/jupyter_notebook/jupyter_submit.sbatch.sh to your directory</li>
| + | |
| − | <li>Modify jupyter_submit.sbatch.sh as needed - instructions are provided in jupyter_submit.sbatch.sh</li>
| + | |
| − | <li>Submit jupyter_submit.sbatch.sh</li>
| + | |
| − | </ul>
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Submit script behaves strangely</font></strong>==
| + | We will now submit the hostname.m function to the cluster. In the Matlab window enter the following: |
| − | ===The statements in my submit script look correct, but it appears that the script is not being read correctly. Why is that ?===
| + | |
| − | This behavior can happen if your submit script contains control characters that confuse
| + | |
| − | the SGE job scheduler. The most common cause of the problem is the presence of Windows | + | |
| − | DOS line feed characters in the submit script. One way to remove these characters is via
| + | |
| − | the Linux <em>dos2unix</em> command. See "man dos2unix".
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Submitting jobs</font></strong>==
| + | <pre>>> j=c.batch(@hostname, 1, {}, 'AutoAddClientPath', false); </pre> |
| − | ===How are jobs submitted to the HPC cluster compute nodes ?===
| + | |
| − | Jobs must be submitted using a "submit script". See [[SonOfGridEngine]].
| + | |
| | | | |
| − | ===What are the valid queue names ?===
| + | @: Submitting a function. <br> |
| − | Valid queue names are : "short", "medium", and "long", and others. See [[KongQueuesTable]] and [[SthenoQueuesTable]].
| + | 1: The number of output arguments from the evaluated function. <br> |
| | + | {}: Cell array of input arguments to the function. In this case empty.<br> |
| | + | AutoAddClientPath', false: The client path is not available on the cluster. |
| | | | |
| − | ===Can I run jobs on a head node ?===
| + | When the job is submitted, you will be prompted for your password. |
| − | Jobs run on an HPC cluster head node that use non-negligible resources will be
| + | |
| − | automatically terminated, with email to that effect sent to the owner of the job.
| + | |
| | | | |
| − | ===How can I run an interactive job on an HPC cluster compute node ?===
| + | [[File:BatchEnterPasswd.png|900px]] |
| − | Use "qlogin" on a head node. See [[KongQueues]].
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Testing if binaries will run</font></strong>==
| + | To wait for the job to finish enter in the Matlab window: |
| − | ===I have binaries, but no source code. How can I tell if the binaries should run on an HPC cluster ?===
| + | |
| − | You can use the <em>library.check</em> (/usr/ucs/bin/library.check) utility, to check for missing
| + | |
| − | libraries an/or GLIBC_* versions. For usage : "library.check -h".
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Using GPUs</font></strong>==
| + | <pre>j.wait</> |
| − | ===What graphical processing units (GPUs) are available in the HPC clusters ?===
| + | |
| − | See [https://web.njit.edu/topics/hpc/specs Cluster specs]
| + | |
| | | | |
| − | ===How do I use those GPUs ?===
| + | Finally, to get the results: |
| − | See [[RunningCUDASamplesOnKong]], [[MatlabGPUOnStheno]]
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Using GPUs in parallel on Kong</font></strong>==
| + | <pre>fetchOutputs(j) |
| − | ===What is the maximum number of GPUs that can be used in parallel on Kong ?===
| + | |
| − | As of April 2016, there are 2 GPU nodes, each with 2 GPUs, so the maximum number
| + | |
| − | of GPUs that can be used in parallel is <strong>4</strong>.
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Using local scratch for large writes and reads</font></strong>==
| + | As can be seen, this job ran on node720 |
| − | ===How do I use the large scratch space local to each compute node to significantlty improve throughput ?===
| + | |
| − | In your submit script, put something like the following :
| + | |
| − | <pre code>
| + | |
| − | ######################################
| + | |
| − | # copy files needed to scratch
| + | |
| − |
| + | |
| − | mkdir -p /scratch/ucid/work
| + | |
| − | cp <all needed files> /scratch/ucid/work
| + | |
| − | cd /scratch/ucid/work
| + | |
| − | #######################################
| + | |
| − | # Run your program
| + | |
| − |
| + | |
| − | /full/path/to/cmd and arguments
| + | |
| − | ########################################
| + | |
| − | # Copy results to local home directory
| + | |
| | | | |
| − | cp <all results and needed files home> ~/results_directory
| + | [[File:BatchHostname.png|900px]] |
| − | #########################################
| + | |
| − | # Delete scratch directory
| + | |
| − |
| + | |
| − | rm -rf /scratch/ucid
| + | |
| − | </pre>
| + | |
| | | | |
| − | ==<strong><font color="#9966cc">Using multithreading</font></strong>==
| |
| − | ===My code uses multithreading. How do I tell the submit script to do multithreading ?===
| |
| − | To use multithreading you need to use the threaded parallel environment. In the submit script :
| |
| − | <pre code>
| |
| − | #$ -pe threaded NUMBER_OF_CORES
| |
| − | </pre>
| |
| | | | |
| − | NUMBER_OF_CORES should not exceed 8 for the short, medium or long queues.
| + | [[File:MonitorJobs.png|900px]] |
| − | | + | ==Submitting a Parallel Function== |
| − | For the <strong>smp</strong> queue, up to 32 cores can be used. In the submit script :
| + | [[File:SubmitParallel.png|900px]] |
| − | <pre code>
| + | [[File:JobFinished.png|900px]] |
| − | #$ -pe threaded 32
| + | [[File:FetchOutputs.png|900px]] |
| − | #$ -q smp
| + | ==Submitting a Script Requiring a GPU== |
| − | </pre>
| + | [[File:GpuSubmitArgs.png|900px]] |
| | + | [[File:GpuSubmit.png|900px]] |
| | + | [[File:GpuDiary.png|900px]] |
| | + | ==Load and Plotting Results from A Job== |
| | + | [[File:PlotDemoSub.png|900px]] |
| | + | [[File:load_x.png|900px]] |
| | + | [[File:plot_x.png|900px]] |
This page is under construction, Will be completed by 6/30
Following this procedure a user will be able to submit jobs to lochness or stheno from Matlab running locally on the user's computer. The version of Matlab on the user's computer must be the same as on the cluster, currently 2021a.
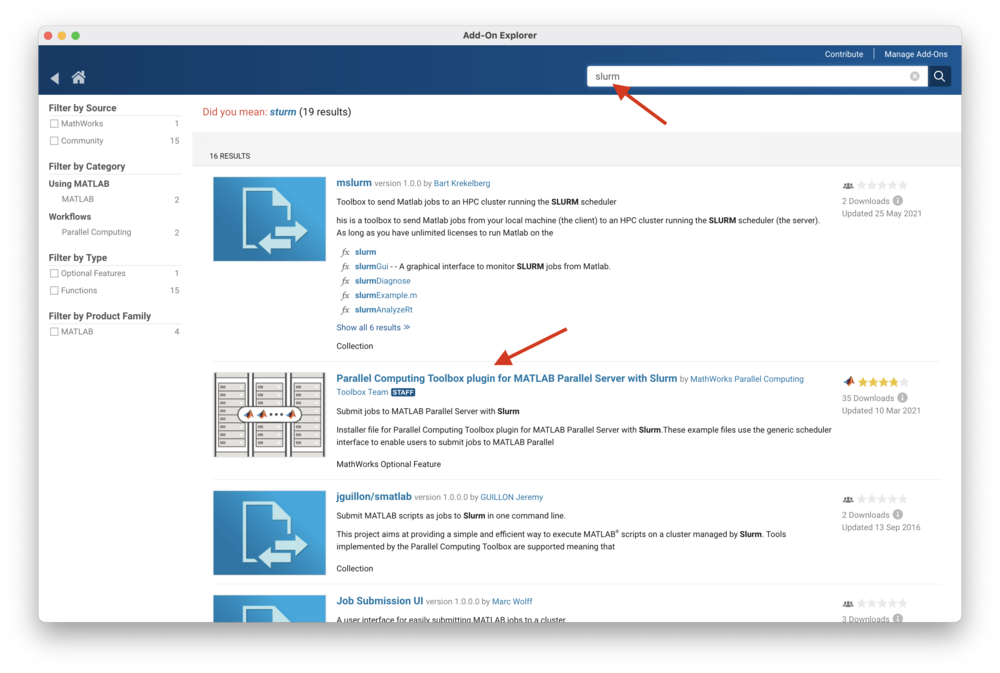
In the search box enter "slurm" and click on the magnifying glass icon.
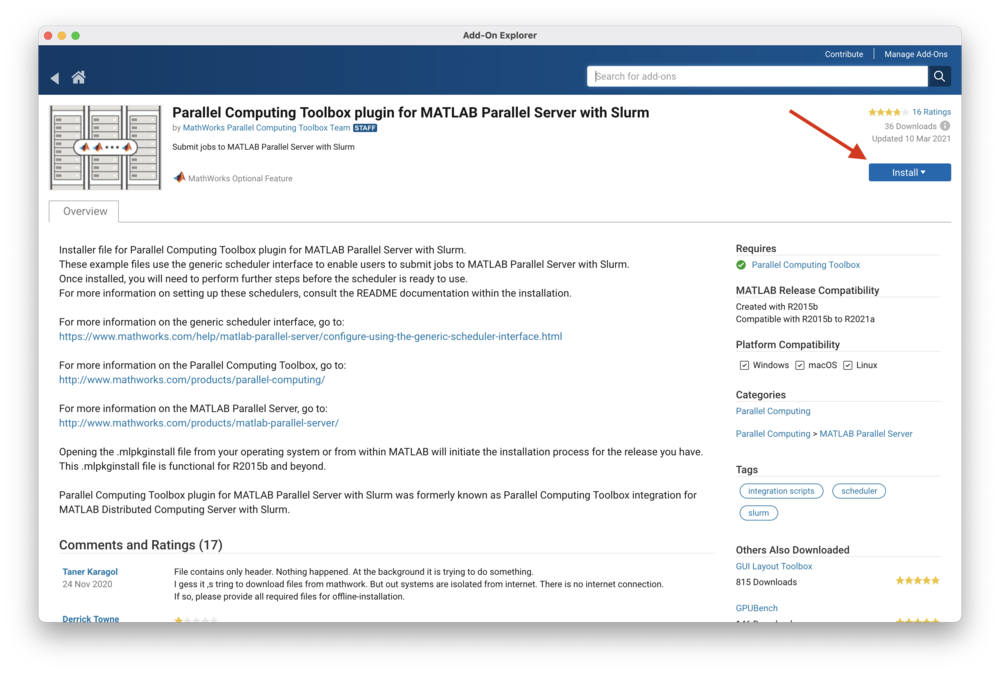
Select "Parallel Computing Toolbox plugin for MATLAB Parallel Server with Slurm"
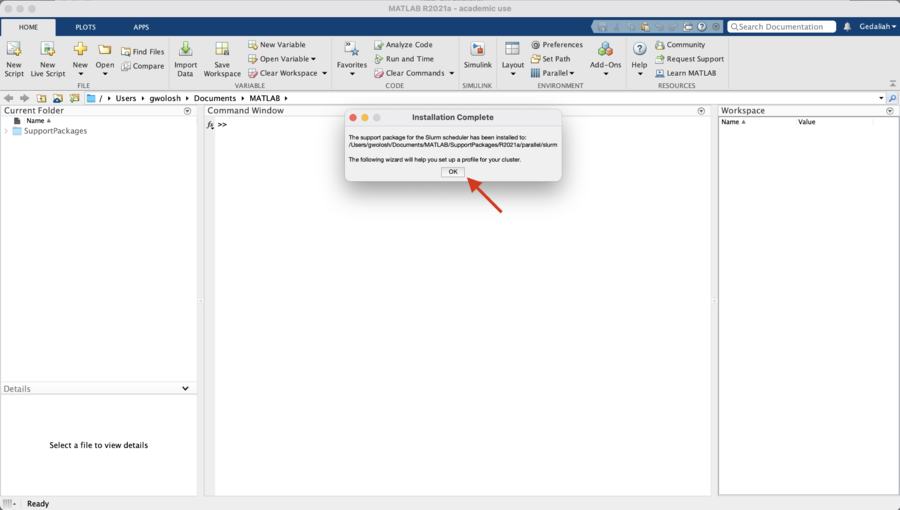
The installation of the Add-On is complete. Click on "OK" the start the "Generic Profile Wizard for Slurm."
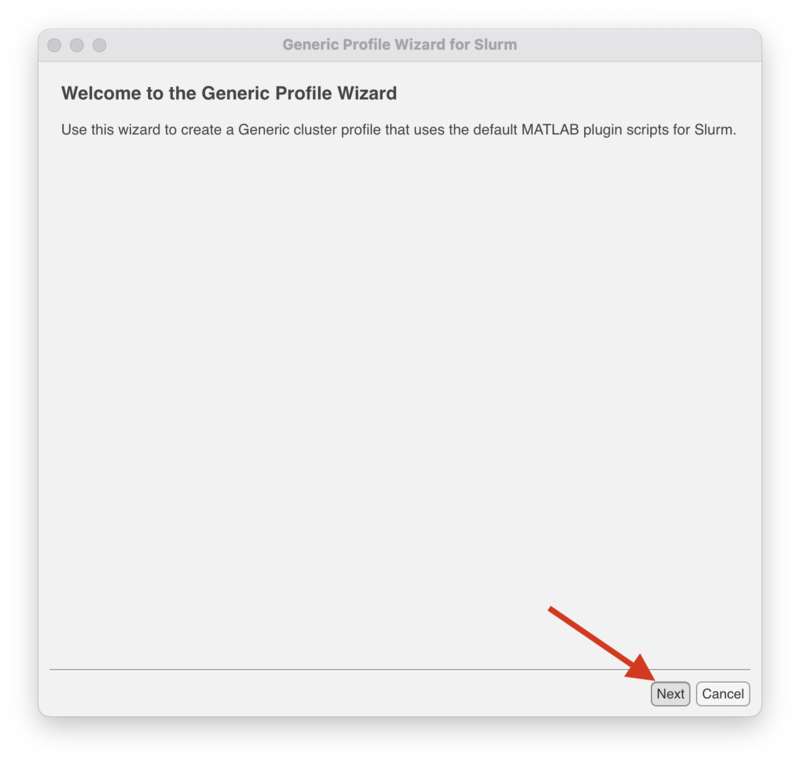
The following steps will create a profile for lochness (or stheno).
Click "Next" to begin.
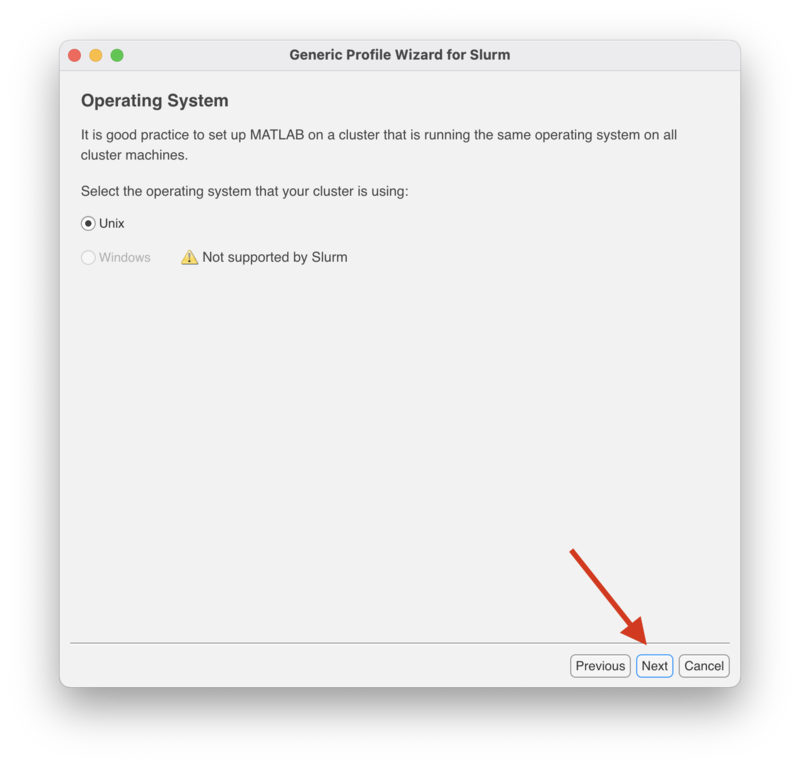
In the "Operating System" screen "Unix" is already selected. Click "Next" to continue.
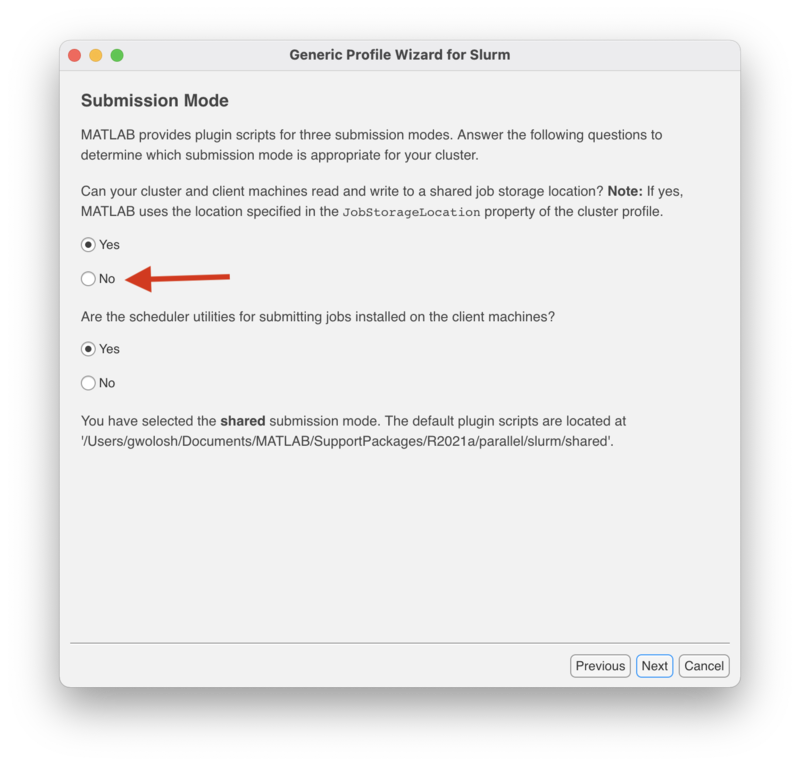
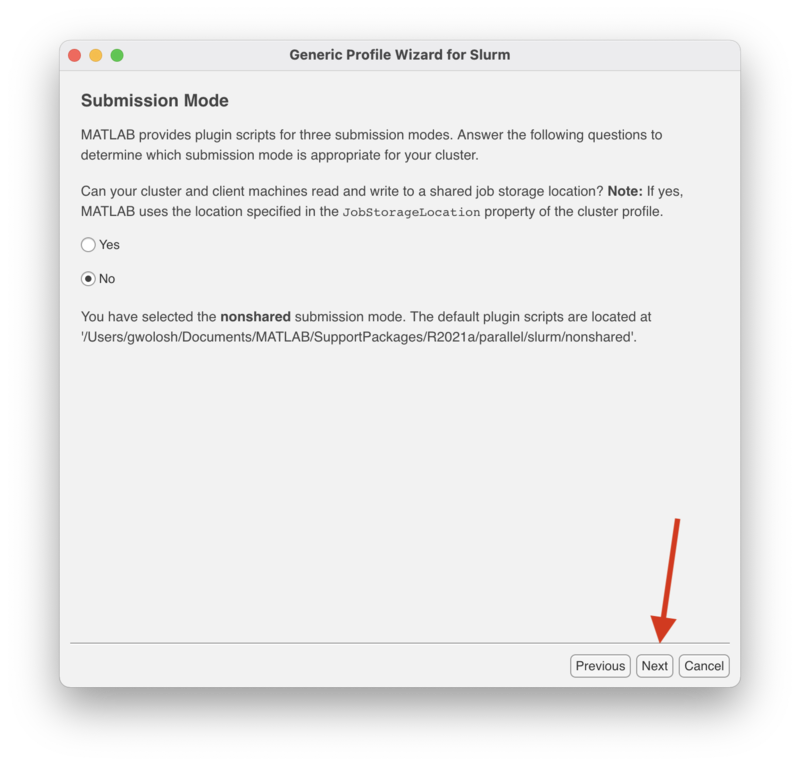
This "Submission Mode" screen determines whether or not to use a "shared" or "nonshared" submission mode. Since Matlab installed on your personal computer or laptop does not use a shared job location storage, select "No" where indicated and click "Next" to continue.
Click "Next" to continue.
In the "Connection Details" screen, enter the cluster host, either "lochness.njit.edu" or "stheno.njit.edu."
Enter your UCID for the username.
Select "No" for the "Do you want to use an identity file to log in to the cluster" option and click next to continue.
In the "Cluster Details" screen enter the full path to the directory on lochness to store the Matlab job files. In the case the directory is $HOME/MDCS. MDCS stands for Matlab Distributed Computing Server. It is not necessary to name this directory MDCS. This directory can be named anything you wish. To determine the value of $HOME, log onto lochness and run the following:
Click "Next" to continue.
In the "Workers" screen enter "512" for the number of workers and "/opt/site/apps/matlab/R2021a" for "MATLAB installation folders for workers."
Click "Next" to continue.
In the "License" screen make sure to select "Network license manager" and click "Next" to continue.
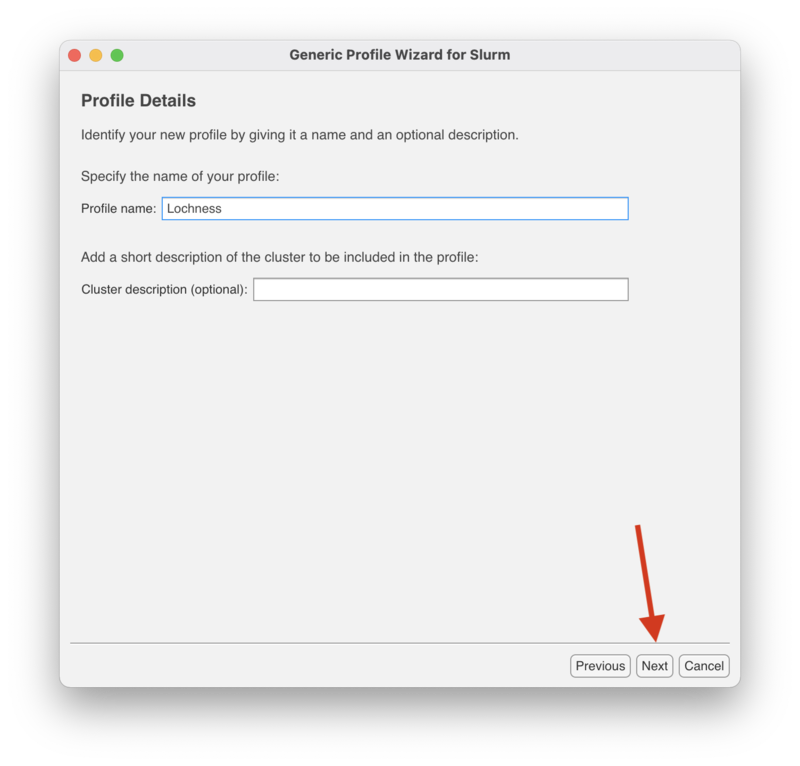
In the "Profile Details" screen enter either "Lochness" or "Stheno" depending on which cluster you are making a profile for. The "Cluster description" is optional and may be left blank.
Click "Next" to continue.
In the "Profile Created Successfully" screen, check the "Set the new profile as default" box and click on "Finish."
This section will demonstrate how to create a cluster object and submit a simple job to the cluster. The job will run the 'hostname' command on the node assigned to the job. The output will indicate clearly that the job ran on the cluster and not on the local computer.
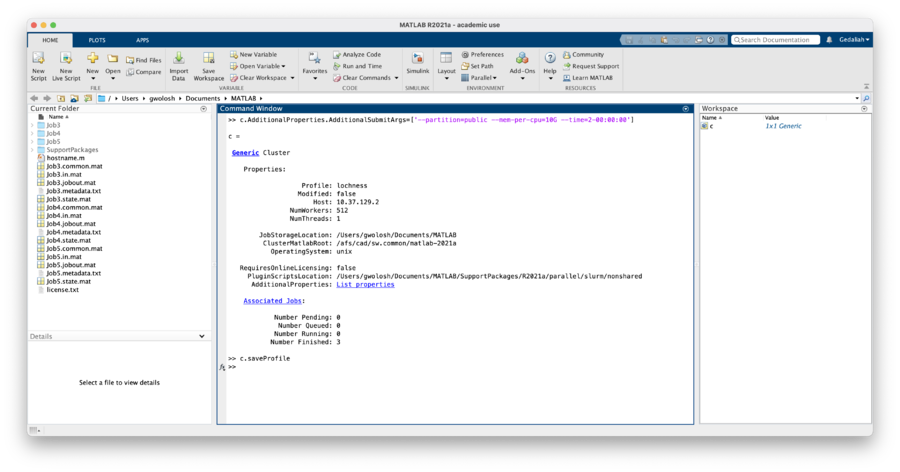
Certain arguments need to be passe dto SLURM in order for the job to run properly. Her we will set values for partion, mem-per-cpu and time. In the Matlab window enter:
To make this persistent between Matlab sessions these arguments need to be saved to the profile. In the Matlab window enter:
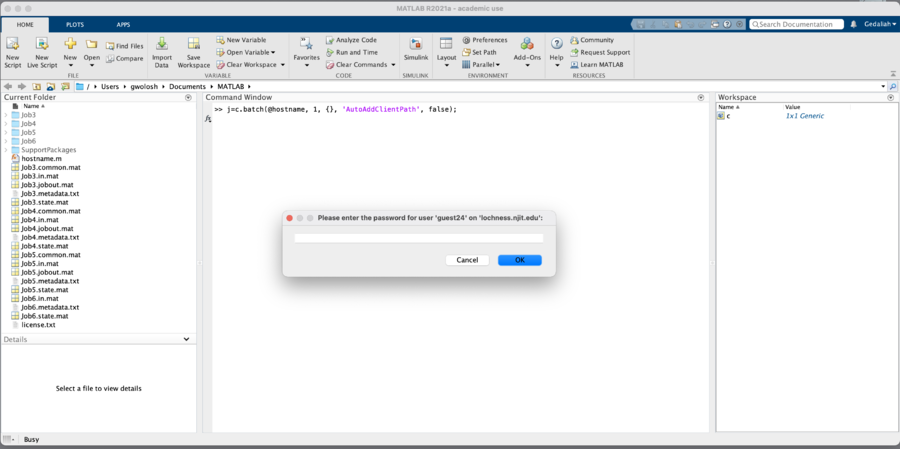
We will now submit the hostname.m function to the cluster. In the Matlab window enter the following:
@: Submitting a function.
1: The number of output arguments from the evaluated function.
{}: Cell array of input arguments to the function. In this case empty.
AutoAddClientPath', false: The client path is not available on the cluster.
When the job is submitted, you will be prompted for your password.